Tailoring DNA Scissors with Engineered High Specificity Glycosylase Enzymes
DNA is charged with carrying life’s secrets though the course of cell division and the lifespan of multicellular organisms. Under frequent attack from environmental hazards, genomes have evolved molecular strategies to maintain integrity via repair systems. Amongst them, DNA glycosylases remove altered base structures, but often have a relaxed substrate specificity. This limits biotechnological applications for precision locating, cutting and mapping of non-canonical structures in mechanistic studies and molecular diagnostics. We are pursuing directed evolution to increase substrate specificity towards specific altered nucleobase structures, and therefore generate novel glycosylases for diverse biotechnological applications
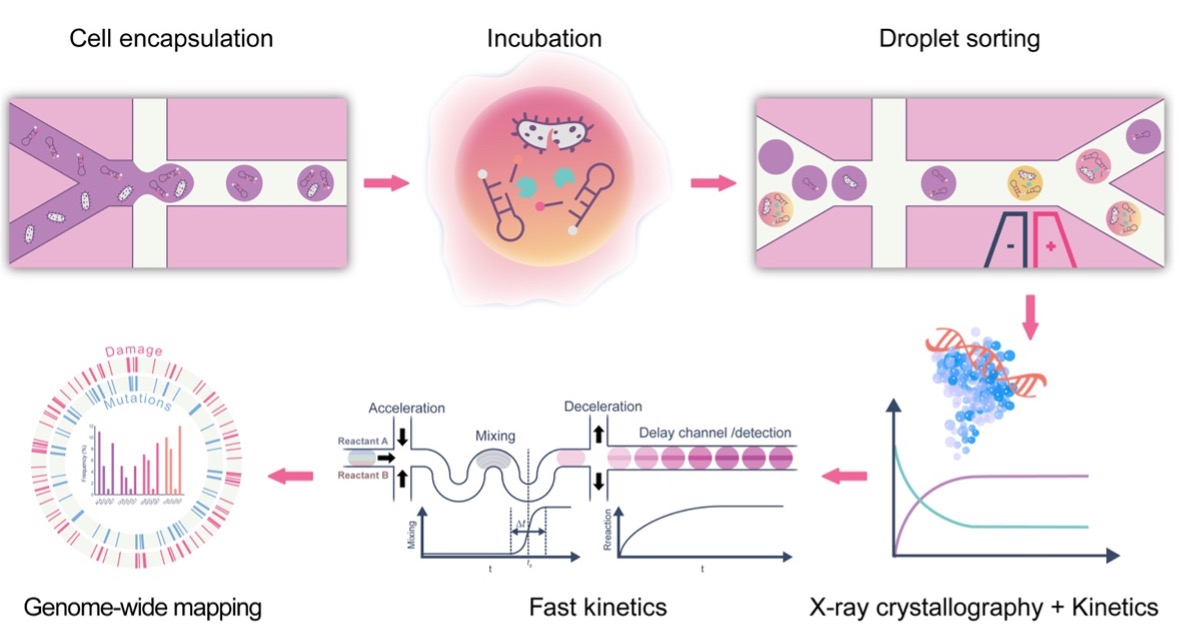
Directed evolution mimics the process of natural selection to evolve tailored proteins. While the capacity to screen very large numbers (ideally >106) of enzyme variants is essential for success, traditional platforms, based on robotic and multi-well plate technologies, are lower throughput than required (allowing between 103 and 105 screens per day) and do not meet the demands of enzyme specificity optimization. Furthermore, current stopped-flow techniques are not ideal for studying the fast kinetics and thermodynamics of thousands of glycosylase enzymes interacting with DNA. To this end, we aim to create an entirely novel and ultra-high-throughput platform for the semi-rational design and directed evolution of glycosylase enzymes as well as the detailed analysis of enzyme variants on ultra-short timescales. Once developed, these enzymes will be utilized to generate high-specificity genomic damage location maps.
This project is supported by the ETH Research Commission and is performed in collaboration with the deMello group (Dr. Stavros Stavrakis and Prof. Dr. Andrew DeMello, ETH Zürich).